TractoR, the medical image analysis platform for which I am the main developer and maintainer, is built on the multiplatform R language, but much of its additional infrastructure is Unix-based. That has meant that researchers using Windows have previously had to jump through some rather awkward hoops to use it.
The newly-released Windows 10 “Anniversary Update” offers an intriguing alternative: an official Linux subsystem, available through a partnership between Microsoft and Canonical, who are behind the popular Ubuntu Linux distribution. Naturally, I had to see whether this would make running TractoR on Windows easier!
Installing the Windows Subsystem for Linux (WSL) is pretty straightforward, as long as you have an up-to-date, and 64-bit, version of Windows 10. After enabling Windows’ developer mode, WSL can be selected from the list of optional Windows features (search from the Start menu). Note that it is tagged as a beta, so there may be issues to be found.
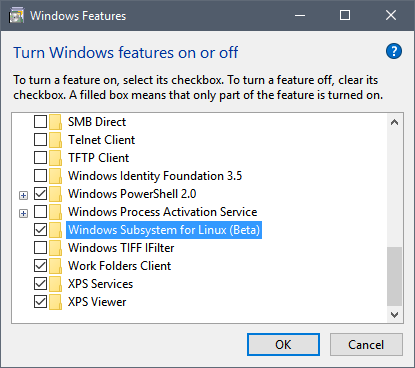
The next step is to open a command prompt, and type bash. This will download the Linux filesystem image from Canonical, install it, and prompt you to create a Unix user. All very simple. Thereafter, you are dropped into a bona fide bash shell, with access to standard Ubuntu commands.
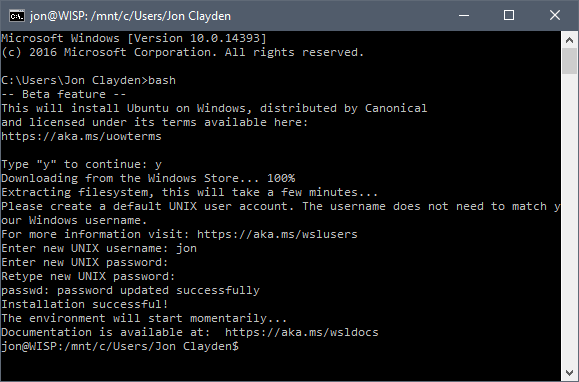
This is when things get interesting. TractoR is based on R, so I needed to install R first of all. On an ordinary Ubuntu system I would type
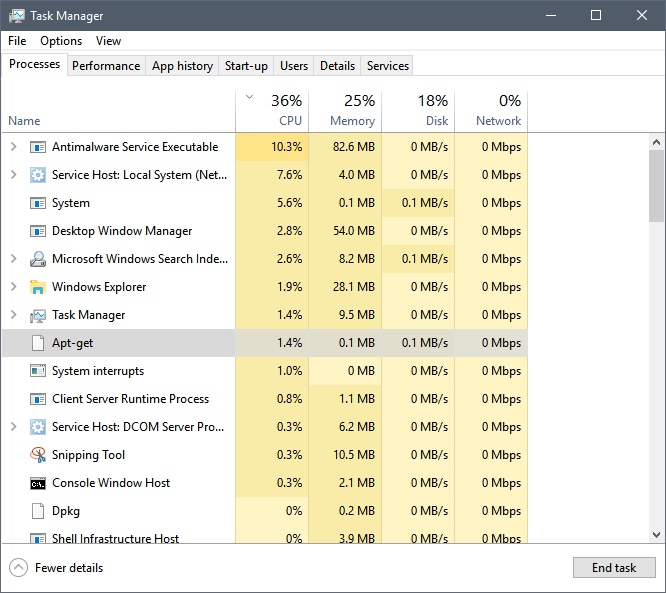
sudo apt-get install r-base-devwhich I did. And, lo and behold, Ubuntu on Windows did as it was told without a hitch. From a geeky point of view it was particularly interesting to note that while apt-get was running, it showed up in the Windows task manager like any native Windows process.
With R installed, I fetched the latest version of TractoR, unzipped it into ~/tractor—a subdirectory of my Linux home directory, not my Windows one—and ran the usual make install. All was well.
The crucial test was... the tests. TractoR comes with a series of self-tests which are designed to ensure that everything is working properly. After setting up the environment, I ran these checks with make test. Apart from a tiny precision mismatch, all went well.
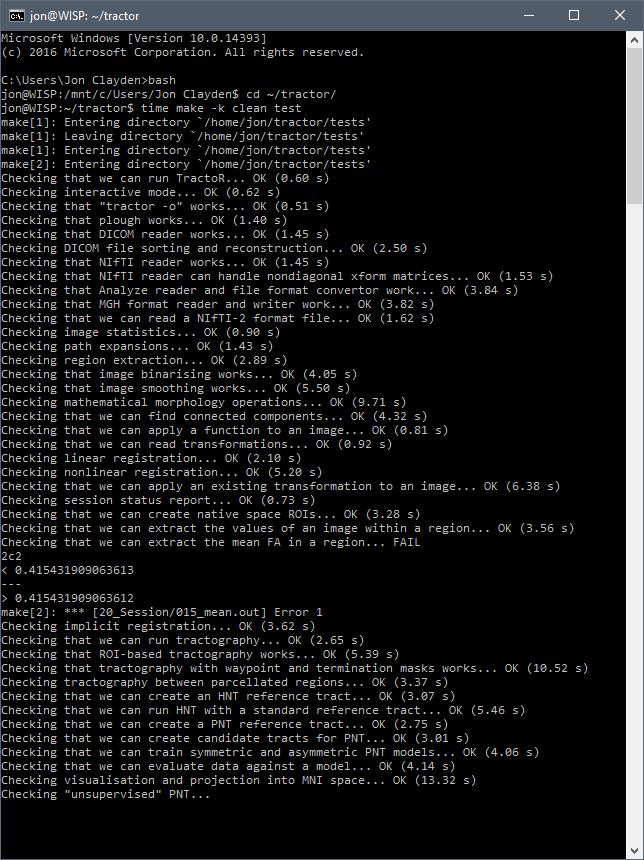
The tests took 3 min 58 sec, a very respectable performance against the 3 min 14 sec the same tests took within macOS on the same machine. So TractoR works fine with WSL, and although there is some virtualisation overhead, it is far from crippling.
The only thing left to do was to check TractoR's graphical capabilities, which require an X window system on Linux. For that I needed a separate X server aimed specifically at Windows—namely (in my case) Xming. After installing the latter package, starting it up and returning to the bash prompt, I could configure the X display using the command
export DISPLAY=localhost:0.0and then try out TractoR's image viewer.
tractor view tests/data/session@refT1 tests/data/session@structural/parcellation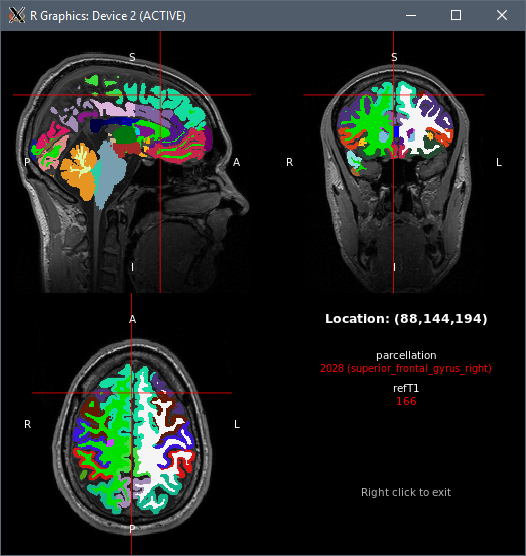
Success! And all within an hour or two of installing the Anniversary Update.
I hope this will make it significantly easier for other researchers to install and run, not just TractoR, but lots of other scientific software on Windows in the future. If you try it, I'd be very glad to hear how you get on!